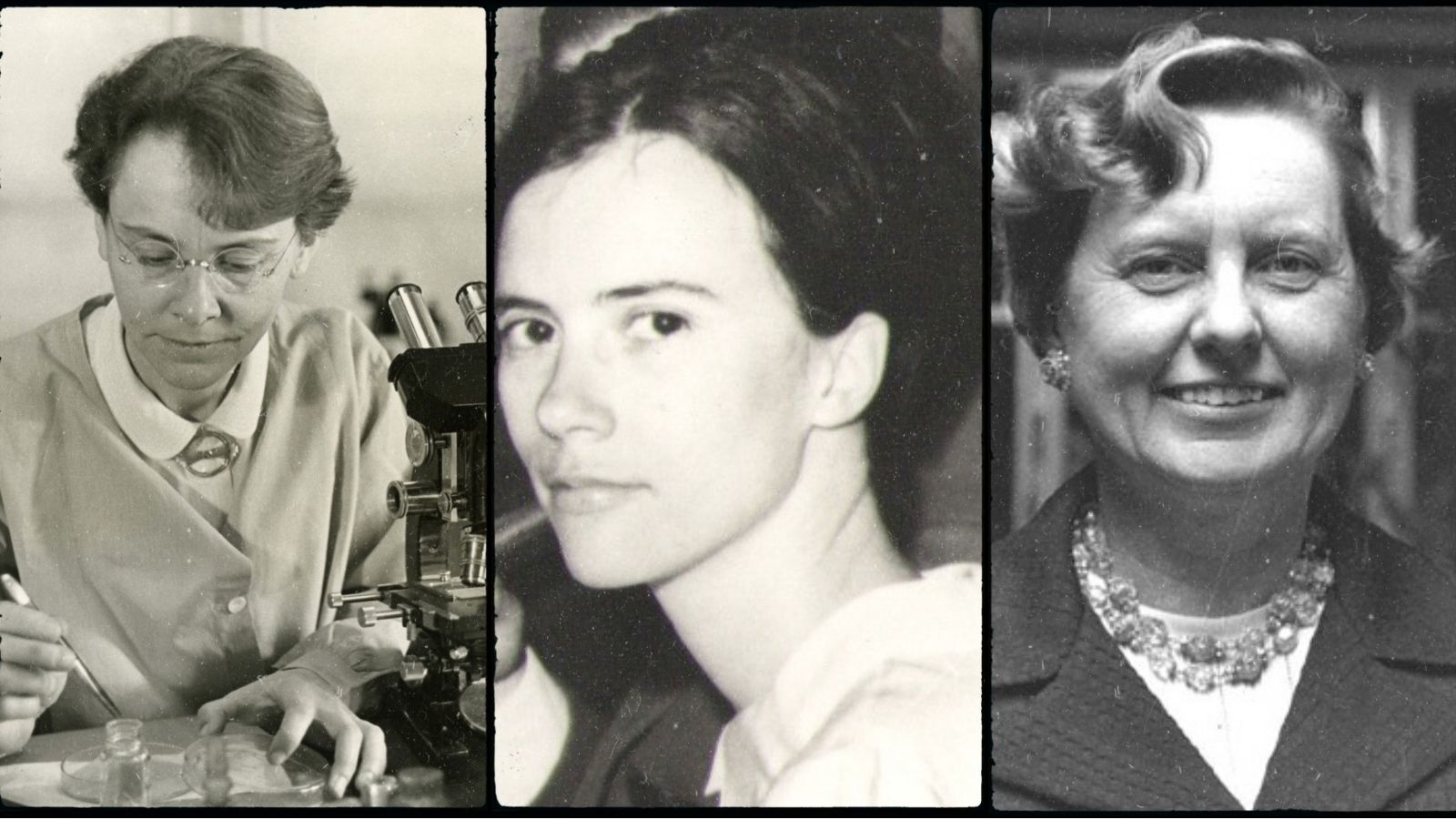
The field of synthetic biology has come a long way from the CRISPR-less days of the early aughts.https://www.nature.com/articles/nrmicro3239 In 2000, bioengineers made the leap from studying natural systems to building the first biological circuits. But it’s common to overlook many of the women in science who laid the groundwork that makes today’s industry possible.Some names are familiar, like Rosalind Franklin, who captured an image of DNA that clearly showed it as a double helix, and Linus Pauling, who illuminated many structural components of proteins. Scientific progress is a massive undertaking that relies on the work of innumerable hardworking researchers whose names don’t always make the history books. In honor of International Women’s Day, here are three overlooked scientists whose research provided key insights into the quirks of genetics, the uses of proteins, and the value of computation in molecular biology. Their work was controversial and undervalued when it was first published, but now it’s clear that these women blazed trails for modern synthetic biology.

Barbara McClintock at work studying genetic crossovers during meiosis. Images: Smithsonian Institution/Science Service; Restored by Adam Cuerden, WikimediaBarbara McClintock came to genetics by way of botany. Her work in maize—a Central American relative of corn—revealed the surprising ways that DNA can move around the chromosome. Her discovery of genetic elements that could move themselves around the genome in a cut-and-paste fashion sparked the most controversy, but after 30 years, it earned her the Nobel prize.After McClintock completed her Ph.D. at Cornell University in 1927, she studied the ten maize chromosomes. In 1931, she demonstrated the physical process of crossing-over, where pairs of chromosomes swap chunks of material before division in meiosis.https://www.pnas.org/content/109/50/20198 By the 1940s, she was a leading scientist in genetics, becoming the third woman in the National Academy of Sciences and the first woman elected president of the Genetics Society of America.https://profiles.nlm.nih.gov/spotlight/ll/feature/biographicalThen, in 1950, she published her first paper about transposons, genetic elements that could remove themselves from the chromosome and insert themselves somewhere else. Geneticist Evelyn Witkin, who was in the audience when McClintock first presented her results, recalled the talk was met with absolute silence.https://www.britannica.com/biography/Evelyn-M-Witkin McClintock herself characterized the response as “puzzlement, even hostility,” because her results challenged the status quo of genetics research at the time.
Chromosomes cross over during the miosis stage of cell division, enabling the chromosomes to swap genetic material. This swap was a critical discovery by Barabara McClintock via GfycatTransposons did not gain widespread recognition until the 1970s when scientists discovered them in bacteria and fruit flies. A research team at the Carnegie Institution for Science isolated and cloned the specific system that McClintock studied, called Ac and Ds, in 1983.https://www.sciencedirect.com/science/article/abs/pii/009286748390226X#! Only then did McClintock receive the Nobel Prize for her discovery of mobile genetic elements. Now there are dozens of transposon-based tools for synthetic biology.http://bergmanlab.genetics.uga.edu/?page_id=295 https://www.frontiersin.org/articles/10.3389/fbioe.2014.00046/full Since they pack their own proteins for snipping DNA out of the genome and splicing it in somewhere else, transposons have become a versatile, standard part of the synthetic biology toolbox.

Daisy Roulland-Dussoix and an illustration showing the restriction site for the restriction enzyme EcoRI.In 1978, the Nobel Prize in Medicine was awarded to three men for their study of restriction enzymes and their applications. Werner Arber received credit for discovering the proteins that bacteria use as a defense against phage because they snip through DNA at specific, symmetrical nucleotide sequences. His collaborator, Daisy Roulland-Dussoix, was left off the prize.Roulland-Dussoix worked with Arber while completing her Ph.D. at the University of Geneva. In two papers published in 1962, Roulland-Dussoix (then Dussoix) and Arber laid the groundwork for the study of restriction enzymes. https://www.sciencedirect.com/science/article/abs/pii/S002228366280059X?via%3Dihub The papers detailed the way that bacteriophages—viruses that infect bacteria—can only infect certain strains. If they do try to infect other strains, the bacteria can somehow recognize and degrade their DNA. Later research revealed that restriction enzymes did that degradation."I am very furious, because apparently he has not even mentioned my name, and I have done half of the work for which he received the Nobel Prize,” wrote Roulland-Dussoix to her brother and sister-in-law in 1978, shortly after the prize was announced.https://upload.wikimedia.org/wikipedia/commons/1/1c/Lettres_Daisy_Dussoix_1978.pdf She wrote that Arber forced her to change her research focus from restriction to radiation in the last year of her studies. During that time, he completed additional research isolating and identifying the enzymes.https://www.cshlpress.com/pdf/RestrictionEnzymes.pdf“In any event,” Roulland-Dussoix wrote, “I think that the Nobel Prize was awarded for the two papers published in 62.”In 1968, Roulland-Dussoix moved to the University of California, San Francisco, where she studied proto-oncogenes (genes with the potential to cause cancer if they mutate) with Harold E. Varmus. He was awarded the Nobel Prize in 1989. Once again Roulland-Dussoix’s name was left out. Roulland-Dussoix did not receive recognition from Arber for her contributions to the Nobel-winning work on restriction enzymes during her lifetime. Now, bioengineers rely on libraries of restriction enzymes for vital tasks like piecing together plasmids thanks to the predictable sticky ends that they leave at cutting sites.

Margaret Dayhoff (Photo credit: Ruth Dayhoff) and the evolutionary tree of human mitogen-activated protein kinases (MAPKs)Margaret Dayhoff’s appreciation for the power of computing in molecular biology led her to invent the field of bioinformatics.https://www.genome.gov/genetics-glossary/Bioinformatics While she used an IBM 7090 mainframe to analyze peptide sequences in the 1960s, many biochemists still dismissed statistical analysis and computing as "theory."Dayhoff began her career at Columbia University in the 1940s. She used punched-card computers to calculate the resonant energies of certain organic compounds and published her findings in 1949. She then took a few years away from the lab to raise her two daughters. When she returned to work in 1962, the National Institutes of Health rejected a grant application on the basis that she “ been out of really intimate touch for some time … with this complicated and rapidly advancing area,” historian Bruno Strassern wrote in Collecting Experiments: Making Big Data Biology. Dayhoff soon joined Robert Ledley at the National Biomedical Research Foundation in Silver Spring, Maryland, where she began cataloging the structures of proteins shared by many species. Previous research had tried to compare many species’ insulin structures, for example, to try to find the most important and functional regions.https://www.jstor.org/stable/40984880?seq=1 But Dayhoff, working with Ledley and Richard Eck, had another goal: to centralize all known protein structures into one publication. The result, the Atlas of Protein Sequence and Structure, was published in 1965. The Atlas offered an open-ended resource for studying molecular biology, and it was even accompanied by possibly the first computerized collection of data in the life sciences. Dayhoff showed that these catalogs could be used to map out evolutionary trees based on the similarities and differences between species’ versions of proteins. Along the way, she invented the one-letter shorthand for amino acids as a way to save space in data storage. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5037978/ http://130.88.97.239/bioactivity/aacodefrm.html“Collecting, comparing, and classifying things of nature seemed old-fashioned to many experimental biologists in the second half of the 20th century,” said Strasser to Smithsonian magazine’s Leila McNeill. But today, “every single publication in experimental biology contains a combination of new experimental data and inferences drawn from comparisons with other data made available in a public database, an approach that Dayhoff started half a century ago.”These pioneers were frequently overlooked and even scorned in their lifetimes. But they played a major part in making the discoveries and building the tools that laid the foundation for modern biology. Today, they are recognized for their groundbreaking achievements in the field now known as synthetic biology.